Method Development for Microbial Ecology
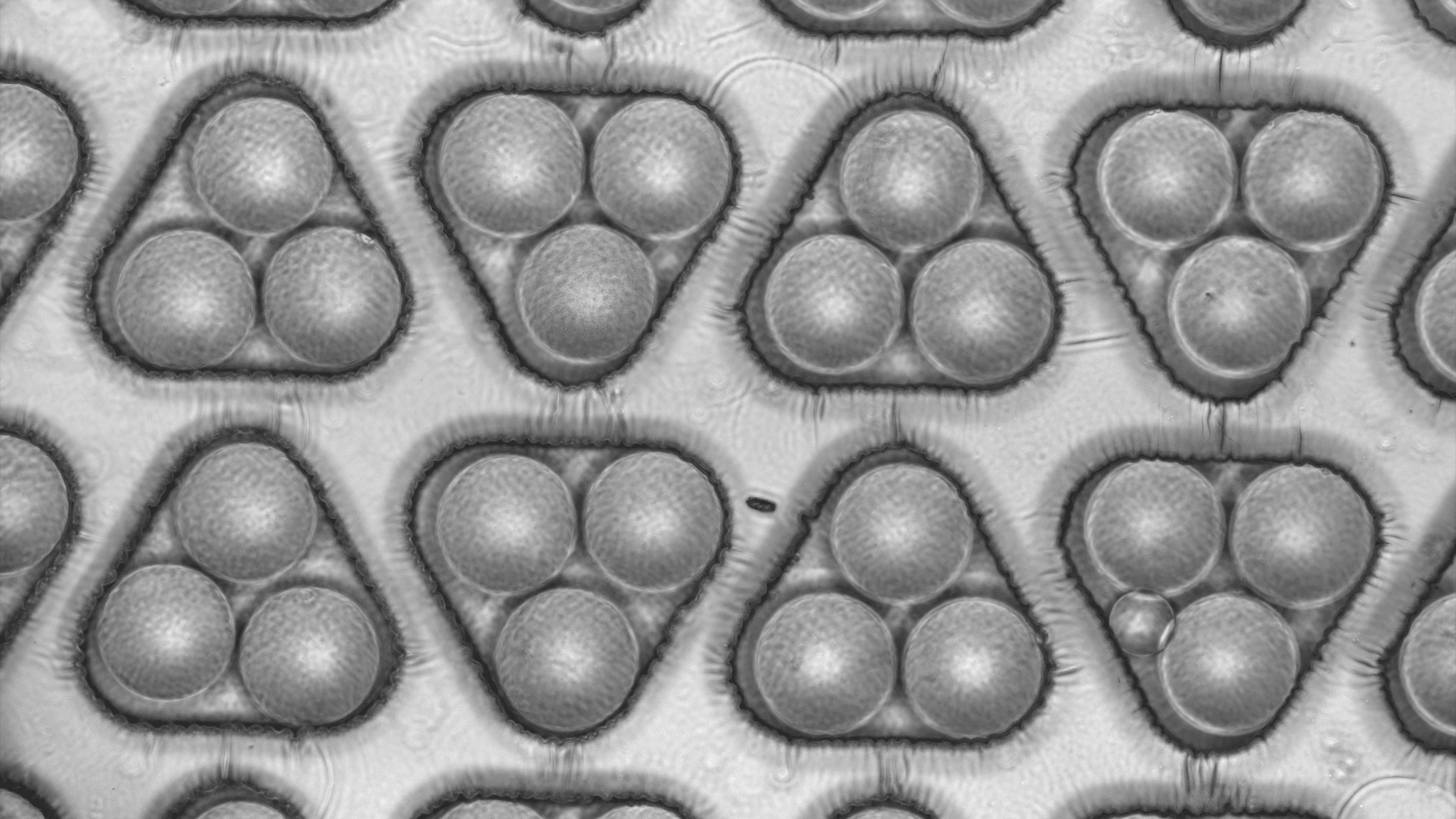
Close-up view of a kChip filled with droplets in which growth of bacterial cells can be monitored at high-throughtput.
Developing and implementing new methods enables us to overcome existing limitations and obtain more accurate, comprehensive data. We are mostly working on methods, tools and software related to microfluidics and live-cell imaging, inluding:
- external page Midap, an automated analysis software for live-cell imaging data (with Franziska Oschmann)
- Microfluidic designs to study microbial interactions (with Stefano Ugolini)
- A combined microfluidics and nanoSIMS method to couple single-cell growth with metabolic activity (Glen, Benjamin, Astrid, external page Fabai Wu, external page Victoria Orphan group)
- A device to perform anaerobic microfluidics-based live-cell imaging (with external page Alyson Hockenberry, Markus Arnoldini and external page Susan Schlegel)
- The kChip, a droplet-based platform designed for the rapid, massively parallel construction and screening of synthetic microbial communities (Anna, originally developed by the external page Blainey lab)